- #1
- 3,759
- 4,199
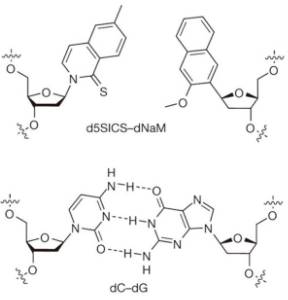
Published today in Nature, researchers report engineering an organism capable of replicating DNA with an unnatural base pair (X-Y) in addition to the two normal base pairs found in DNA (A-T and C-G).
Here's what the unnatural base pair looks like (in comparison to the C-G base pair):
Here's the abstract for the article:
Malyshev et al. A semi-synthetic organism with an expanded genetic alphabet. Nature. Published online 07 May 2014. http://dx.doi.org/10.1038/nature13314 .
It'll be interesting to see what (if any) applications come from this work. It seems like there's still some work to be done in order to make it a little easier for these organisms to naturally use the new bases (for example, maybe getting the organisms to synthesize them by themselves).
Here's what the unnatural base pair looks like (in comparison to the C-G base pair):
Here's the abstract for the article:
Organisms are defined by the information encoded in their genomes, and since the origin of life this information has been encoded using a two-base-pair genetic alphabet (A–T and G–C). In vitro, the alphabet has been expanded to include several unnatural base pairs (UBPs). We have developed a class of UBPs formed between nucleotides bearing hydrophobic nucleobases, exemplified by the pair formed between d5SICS and dNaM (d5SICS–dNaM), which is efficiently PCR-amplified and transcribed in vitro, and whose unique mechanism of replication has been characterized. However, expansion of an organism’s genetic alphabet presents new and unprecedented challenges: the unnatural nucleoside triphosphates must be available inside the cell; endogenous polymerases must be able to use the unnatural triphosphates to faithfully replicate DNA containing the UBP within the complex cellular milieu; and finally, the UBP must be stable in the presence of pathways that maintain the integrity of DNA. Here we show that an exogenously expressed algal nucleotide triphosphate transporter efficiently imports the triphosphates of both d5SICS and dNaM (d5SICSTP and dNaMTP) into Escherichia coli, and that the endogenous replication machinery uses them to accurately replicate a plasmid containing d5SICS–dNaM. Neither the presence of the unnatural triphosphates nor the replication of the UBP introduces a notable growth burden. Lastly, we find that the UBP is not efficiently excised by DNA repair pathways. Thus, the resulting bacterium is the first organism to propagate stably an expanded genetic alphabet.
Malyshev et al. A semi-synthetic organism with an expanded genetic alphabet. Nature. Published online 07 May 2014. http://dx.doi.org/10.1038/nature13314 .
It'll be interesting to see what (if any) applications come from this work. It seems like there's still some work to be done in order to make it a little easier for these organisms to naturally use the new bases (for example, maybe getting the organisms to synthesize them by themselves).
Last edited by a moderator: