- #1
- 2,355
- 10
It is often useful to be able to compute homology and cohomology without too much thought. Macaulay2 is an outstanding example of a open source software project under active development by mathematicians
http://www.math.uiuc.edu/Macaulay2/
Macaulay2 was created primarily to do the kind of homological algebra which is useful in algebraic geometry, and the help pages (which are extensive, although some important utilities remain undocumented) will make much more sense if you have studied a textbook which takes a very similar viewpoint:
Eisenbud, Commutative Algebra with a Viewpoint Towards Algebraic Geometry, Springer,GTM 150
There is even a very nice textbook which uses Macaulay2:
Schenck, Computational Algebraic Geometry, Cambridge University Press, LMS student texts 58.
These are both beautiful books and I recommend them both (the second is much easier), but in this thread a more important reference will be a third lovely textbook
Hatcher, Algebraic Topology, Cambridge University Press
which is available for free download at Hatcher's academic website.
Some more standard references, which you can use to check my computations:
May, A Concise Course in Algebraic Topology, University of Chicago Press, 1999
Munkres, Elements of Algebraic Topology, Benjamin/Cummings, 1984
Greenberg & Harper, Algebraic Topology: A First Course, Addison-Wesley, 2nd ed, 1981
Hilton & Wylie, Homology Theory, Cambridge University Press, 1960
To save time and energy, I'll assume--- at least until further notice--- that the reader has installed Macaulay2 and is using it with emacs (chain complexes tend to be rather long and in emacs we can scroll horizontally, which can be very helpful in keeping output legible).
[size=+1]The Circle[/size]
Let's start with something really simple. Suppose we have a circle, which we have represented as a simplicial complex using three edges. We naturally want to think of the boundary map from one-chains to zero-chains as a matrix:
[tex]
\partial_1 =
\left[ \begin{array}{rrr|l}
e_1 & e^\prime_1 & e^{\prime \prime}_1 & \\
\hline
1 & 0 & 1 & e_0 \\
-1 & 1 & 0 & e^\prime_0 \\
0 & -1 & -1 & e^{\prime \prime}_0
\end{array} \right]
[/tex]
where I have labled the columns by the three one-cells and the rows by the three zero-cells. That is, [itex]\partial_1 e_1 = e_0 - e^\prime_0[/itex] and so on, which tells what picture I have in mind and also how the cells are labeled.
Thus, our chain complex
[tex]
0 \leftarrow C^0(X,\mathbb{Z}) \stackrel{\partial_1}{\leftarrow} C^1(X,\mathbb{Z}) \leftarrow 0
[/tex]
will be
[tex]
0 \leftarrow \mathbb{Z}^3
\stackrel{
{\small \left[ \begin{array}{rrr}
1 & 0 & 1 \\
-1 & 1 & 0 \\
0 & -1 & -1
\end{array} \right] }
}{\longleftarrow}
\mathbb{Z}^3 \leftarrow 0
[/tex]
and we wish to compute the homology. (If we use left arrows, matrix multiplication of the matrix times a column vector of coefficients will correspond to evaluating the boundary of some chain.)
Here's how we do this in Macaulay2: call emacs and hit the F12 key to invoke Macaulay2:
This means that the homology sequence
[tex]
0
\leftarrow H_0(X,\mathbb{Z})
\stackrel{\partial_1}{\leftarrow} H_1(X,\mathbb{Z})
\leftarrow 0
[/tex]
is simply
[tex]
0
\leftarrow \mathbb{Z}
\stackrel{\partial_1}{\leftarrow} \mathbb{Z}
\leftarrow 0
[/tex]
which means (since [itex]H_0 =\mathbb{Z}[/itex]) that we have a connected space and a single generating cycle (since [itex]H_1 =\mathbb{Z}[/itex]).
To see the boundary operator, do this:
Try omitting "prune" to see how Macualay2 reports homology in its full glory.
Did you spot the nifty trick? Macaulay2 knows all about tensor products, so it knows that when we tensor an integer matrix with a ring we get a matrix whose entries are interpreted as ring elements. Since I'm using the ring of integers, this may not seem to matter, but Macaulay2 is very strongly typed, which is actually one of the really nice things about using it--- Macaulay2 insists at every step that you know whether the thing you are computing is a ring element in a particular ring, a graded module over a particular ring, or whatnot.
Something I plan to sweep under the rug (but see Eisenbud) is that at this level of structure, there is virtually no distinction between homology and cohomology. Of course we know that in algebraic topology, the cohomology module (a graded module) acquires a ring structure (making it a graded ring) via the cup product. We are often interested in working with this ring structure; Macaulay2 does offer specialized packages which can do this for, in particular, the cohomology of complex Grassmann manifolds (the space of projective k-flats in some projective space).
I'm tacitly assuming that the reader has some prior exposure to homology, but to prevent any possible misunderstanding, let me stress than the chain groups (and the groups of cycles and of boundaries) are not topological invariants; only the sequence of homology groups is a topological invariant. In homological algebra, we consider the sequence of homology groups (these are always abelian groups, of course) as a single object, a graded module over the integers. Then we generalize from the ring of integers to any ring R, so that we have a graded module over R. It turns out to be useful to be able to compute homology modulo a prime, for example.
Simplicial chain complexes tend to be absurdly bloated, however, and we'd like to cut way down on the extraneous junk in our chain complexes by thinking of a circle as two one-cells attached to two zero-cells. So suppose we "cheat" and use an "illegal" chain complex with boundary map
[tex]
\partial_1 =
\left[ \begin{array}{rr|l}
e_1 & e^\prime_1 & \\
\hline
1 & -1 & e_0 \\
-1 & 1 & e^\prime_0
\end{array} \right]
[/tex]
Then we still get the correct homology sequence!
Suppose we are bolder and try to think of the circle as a single one-cell attached to a single zero-cell. Then the boundary map is trivial
[tex]
\partial_1 =
\left[ \begin{array}{r|l}
e_1 & \\
\hline
0 & e_0
\end{array} \right]
[/tex]
but we still get the correct homology sequence!
But you should be wary--- it turns out that this last simplification took things a bit too far, even for the purposes of "cheating"!
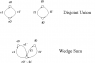
[size=+1]Wedges of Circles[/size]
Now consider three one dimensional spaces which are all homotopy equivalent to a wedge of two circles
http://www.math.uiuc.edu/Macaulay2/
Macaulay2 was created primarily to do the kind of homological algebra which is useful in algebraic geometry, and the help pages (which are extensive, although some important utilities remain undocumented) will make much more sense if you have studied a textbook which takes a very similar viewpoint:
Eisenbud, Commutative Algebra with a Viewpoint Towards Algebraic Geometry, Springer,GTM 150
There is even a very nice textbook which uses Macaulay2:
Schenck, Computational Algebraic Geometry, Cambridge University Press, LMS student texts 58.
These are both beautiful books and I recommend them both (the second is much easier), but in this thread a more important reference will be a third lovely textbook
Hatcher, Algebraic Topology, Cambridge University Press
which is available for free download at Hatcher's academic website.
Some more standard references, which you can use to check my computations:
May, A Concise Course in Algebraic Topology, University of Chicago Press, 1999
Munkres, Elements of Algebraic Topology, Benjamin/Cummings, 1984
Greenberg & Harper, Algebraic Topology: A First Course, Addison-Wesley, 2nd ed, 1981
Hilton & Wylie, Homology Theory, Cambridge University Press, 1960
To save time and energy, I'll assume--- at least until further notice--- that the reader has installed Macaulay2 and is using it with emacs (chain complexes tend to be rather long and in emacs we can scroll horizontally, which can be very helpful in keeping output legible).
[size=+1]The Circle[/size]
Let's start with something really simple. Suppose we have a circle, which we have represented as a simplicial complex using three edges. We naturally want to think of the boundary map from one-chains to zero-chains as a matrix:
[tex]
\partial_1 =
\left[ \begin{array}{rrr|l}
e_1 & e^\prime_1 & e^{\prime \prime}_1 & \\
\hline
1 & 0 & 1 & e_0 \\
-1 & 1 & 0 & e^\prime_0 \\
0 & -1 & -1 & e^{\prime \prime}_0
\end{array} \right]
[/tex]
where I have labled the columns by the three one-cells and the rows by the three zero-cells. That is, [itex]\partial_1 e_1 = e_0 - e^\prime_0[/itex] and so on, which tells what picture I have in mind and also how the cells are labeled.
Thus, our chain complex
[tex]
0 \leftarrow C^0(X,\mathbb{Z}) \stackrel{\partial_1}{\leftarrow} C^1(X,\mathbb{Z}) \leftarrow 0
[/tex]
will be
[tex]
0 \leftarrow \mathbb{Z}^3
\stackrel{
{\small \left[ \begin{array}{rrr}
1 & 0 & 1 \\
-1 & 1 & 0 \\
0 & -1 & -1
\end{array} \right] }
}{\longleftarrow}
\mathbb{Z}^3 \leftarrow 0
[/tex]
and we wish to compute the homology. (If we use left arrows, matrix multiplication of the matrix times a column vector of coefficients will correspond to evaluating the boundary of some chain.)
Here's how we do this in Macaulay2: call emacs and hit the F12 key to invoke Macaulay2:
Code:
Macaulay2, version 1.3.1
with packages: ConwayPolynomials, Elimination, IntegralClosure, LLLBases,
PrimaryDecomposition, ReesAlgebra, SchurRings, TangentCone
i1 : R = ZZ
o1 = ZZ
o1 : Ring
i2 : S1 = matrix{{1,0,1},{-1,1,0},{0,-1,-1}} ** R
o2 = | 1 0 1 |
| -1 1 0 |
| 0 -1 -1 |
3 3
o2 : Matrix ZZ <--- ZZ
i3 : S = chainComplex(S1)
3 3
o3 = ZZ <-- ZZ
0 1
o3 : ChainComplex
i4 : prune HH S
1
o4 = 0 : ZZ
1
1 : ZZ
o4 : GradedModule[tex]
0
\leftarrow H_0(X,\mathbb{Z})
\stackrel{\partial_1}{\leftarrow} H_1(X,\mathbb{Z})
\leftarrow 0
[/tex]
is simply
[tex]
0
\leftarrow \mathbb{Z}
\stackrel{\partial_1}{\leftarrow} \mathbb{Z}
\leftarrow 0
[/tex]
which means (since [itex]H_0 =\mathbb{Z}[/itex]) that we have a connected space and a single generating cycle (since [itex]H_1 =\mathbb{Z}[/itex]).
To see the boundary operator, do this:
Code:
i46 : S.dd
3 3
o46 = 0 : ZZ <---------------- ZZ : 1
| 1 0 1 |
| -1 1 0 |
| 0 -1 -1 |
o46 : ChainComplexMapDid you spot the nifty trick? Macaulay2 knows all about tensor products, so it knows that when we tensor an integer matrix with a ring we get a matrix whose entries are interpreted as ring elements. Since I'm using the ring of integers, this may not seem to matter, but Macaulay2 is very strongly typed, which is actually one of the really nice things about using it--- Macaulay2 insists at every step that you know whether the thing you are computing is a ring element in a particular ring, a graded module over a particular ring, or whatnot.
Something I plan to sweep under the rug (but see Eisenbud) is that at this level of structure, there is virtually no distinction between homology and cohomology. Of course we know that in algebraic topology, the cohomology module (a graded module) acquires a ring structure (making it a graded ring) via the cup product. We are often interested in working with this ring structure; Macaulay2 does offer specialized packages which can do this for, in particular, the cohomology of complex Grassmann manifolds (the space of projective k-flats in some projective space).
I'm tacitly assuming that the reader has some prior exposure to homology, but to prevent any possible misunderstanding, let me stress than the chain groups (and the groups of cycles and of boundaries) are not topological invariants; only the sequence of homology groups is a topological invariant. In homological algebra, we consider the sequence of homology groups (these are always abelian groups, of course) as a single object, a graded module over the integers. Then we generalize from the ring of integers to any ring R, so that we have a graded module over R. It turns out to be useful to be able to compute homology modulo a prime, for example.
Simplicial chain complexes tend to be absurdly bloated, however, and we'd like to cut way down on the extraneous junk in our chain complexes by thinking of a circle as two one-cells attached to two zero-cells. So suppose we "cheat" and use an "illegal" chain complex with boundary map
[tex]
\partial_1 =
\left[ \begin{array}{rr|l}
e_1 & e^\prime_1 & \\
\hline
1 & -1 & e_0 \\
-1 & 1 & e^\prime_0
\end{array} \right]
[/tex]
Then we still get the correct homology sequence!
Suppose we are bolder and try to think of the circle as a single one-cell attached to a single zero-cell. Then the boundary map is trivial
[tex]
\partial_1 =
\left[ \begin{array}{r|l}
e_1 & \\
\hline
0 & e_0
\end{array} \right]
[/tex]
but we still get the correct homology sequence!
Code:
i10 : S1 = matrix{{0}} ** R
o10 = 0
1 1
o10 : Matrix ZZ <--- ZZ
i11 : S = chainComplex(S1)
1 1
o11 = ZZ <-- ZZ
0 1
o11 : ChainComplex
i13 : prune HH S
1
o13 = 0 : ZZ
1
1 : ZZ
o13 : GradedModule[size=+1]Wedges of Circles[/size]
Now consider three one dimensional spaces which are all homotopy equivalent to a wedge of two circles
- two circles joined by an edge
[tex]
\partial_1 =
\left[ \begin{array}{rrr|l}
e_1 & e^\prime_1 & e^{\prime\prime}_1 \\
\hline
1 & 0 & 0 & e_0 \\
-1 & 0 & 0 & e^\prime_0
\end{array} \right]
[/tex] - two circles joined at a common point
[tex]
\partial_1 =
\left[ \begin{array}{rr|l}
e_1 & e^\prime_1 \\
\hline
0 & 0 & e_0
\end{array} \right]
[/tex] - a circle spanned by an edge
[tex]
\partial_1 =
\left[ \begin{array}{rrr|l}
e_1 & e^\prime_1 & e^{\prime\prime}_1 \\
\hline
1 & 1 & 1 & e_0 \\
-1 & -1 & -1 & e^\prime_0
\end{array} \right]
[/tex]
Last edited: