- #1
lavoisier
- 177
- 24
Hello, here's a problem at the boundary between mathematics, programming and chemistry, which I'm struggling with, but I guess can be easily solved by mathematicians/programmers.
I'll explain it as concisely as I can, but sorry, it will be a long post, it's not that trivial.
In chemistry there is an operation called 'R group decomposition' that is performed on a set of molecules to analyse the structural determinants of some of their properties.
Here's an example of template ('scaffold') that can be used:
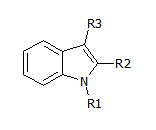
There is some software that takes this template + a set of molecules, checks which ones 'match' the template and lists the corresponding R1, R2 and R3. These are the 'R groups', and taken together with the scaffold, uniquely identify a molecule. So far so good.
However, there are templates that, by effect of symmetry and chemical rules, can produce several valid sets of R groups. For instance this one:
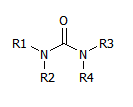
Symmetry shows that R1,R2 can be swapped with R3,R4.
Chemistry shows that R1 and R2 can be swapped, and so can R3 and R4.
So, a molecule like this one:
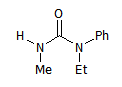 M1
M1
has 8 'mappings':
M1_{R1,R2,R3,R4}={H,Me,Ph,Et},{H,Me,Et,Ph},{Me,H,Ph,Et},{Me,H,Et,Ph},{Ph,Et,H,Me},{Ph,Et,Me,H},{Et,Ph,H,Me},{Et,Ph,Me,H}.
Each of these mappings is perfectly valid. If one were only interested in identifying molecules, choosing anyone mapping at random would be OK.
The problem is that, when analysing the properties of a set of molecules, it is desirable to minimise the number of distinct 'descriptors' at each R-position and maximise the sum of all pairwise differences of numbers of descriptors; choosing mappings at random does not achieve that.
Example: suppose that we choose the first mapping for M1:
M1_{R1,R2,R3,R4}={H,Me,Ph,Et}
In terms of descriptors at each R position, we have: {R1}={H}, {R2}={Me}, {R3}={Ph}, {R4}={Et}.
Now we add this molecule to the set:
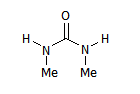 M2
M2
Due to the identity between some groups, the non-degenerate mappings are fewer:
M2_{R1,R2,R3,R4}={H,Me,H,Me},{H,Me,Me,H},{Me,H,Me,H},{Me,H,H,Me}.
It is inevitable to add new descriptors to 'some' R positions.
If we choose the first mapping of M2, we're doing well, because we don't add any new descriptor to R1 or R2, and we're adding new descriptors only to R3 and R4.
M2_{R1,R2,R3,R4}={H,Me,H,Me}
And we have now: {R1}={H}, {R2}={Me}, {R3}={Ph,H}, {R4}={Et,Me}.
The number of descriptors in each R position is just the number of distinct elements in each of these sets (so 1, 1, 2, 2 here). The total number of descriptors is the sum of the above, so here 1+1+2+2=6 (you don't consider equal the H in R1 and the H in R3, because at this point they are desymmetrised.
Obviously if we had chosen the 3rd mapping for M2, we would have added one new descriptor to each position, ignoring the fact that H was already in R1 and Me in R2.
It may seem simple: just choose for each new molecule the mapping that adds the fewest descriptors.
But it's not. First, you don't really add the molecules one by one: you already have a large set of molecules (e.g. 500) with all their combinatorial mappings (could be from 2 to 8, typically, for each molecule), and in the end you need to choose one mapping per molecule in such a way that the above conditions are satisfied (minimal sum of different descriptors, maximal sum of differences of numbers of descriptors). I don't need to tell you how many possible combinations that would make, if one wanted to approach this by brute force :O)
So I went for what I thought was a more 'clever' approach.
I tried this: pick one arbitrary mapping for the first molecule and then run iterations among the rest of the set to choose at each cycle one mapping that gives the lowest addition of descriptors overall, and within this condition, adds the new descriptors preferably to groups that already have more descriptors.
Computationally, I just made a vector like {1,0,0,1} for each mapping, comparing existing descriptors with the one in the mapping, 1 for not present, 0 for already present. Then I made a SUM composed by the norm of the vector (in this case 2) multiplied by 104, + the individual vector components each multiplied by 10i, where i is the appropriately chosen exponent from 0 to 3, based on the sorting by descriptor counts in the current reference sets of descriptors in the R positions. So you have 10001 for a mapping that adds 1 new descriptor to the R position that already has the most descriptors. 10010 would indicate adding 1 new descriptor to the R position that has the second most descriptors, and so on, 20011 would add 1 to each the most populated R position and the second most populated one. So at each cycle only mappings with the lowest SUM are considered, and only one of them is selected (because two mappings with SUM 10001 don't necessarily add the same descriptor to that position). The corresponding molecule passes, the reference R position sets of descriptors are updated, all other molecules are sent to the next iteration.
This seemed to work to some extent, but by running some tests I found that the initial choice, and even the order of the molecules, resulted in different end sets, some with a good separation of descriptors, some less good, and even the total number of descriptors varied a bit.
Example: in one run I got something like 13, 80, 220, 262; in another run something like 40,65,250,230.
This is not good. The solution should not only be unique, but also optimal. I'm not sure it can be independent from the initial choice of mapping for the first compound, but then there should be a way to choose that best.
Here's an example set:
M1 ={H,Me,Ph,Et},{H,Me,Et,Ph},{Me,H,Ph,Et},{Me,H,Et,Ph},{Ph,Et,H,Me},{Ph,Et,Me,H},{Et,Ph,H,Me},{Et,Ph,Me,H}
M2 ={H,Me,H,Me},{H,Me,Me,H},{Me,H,Me,H},{Me,H,H,Me}
M3={H,H,Me,Me},{Me,Me,H,H}
Can anyone please suggest an approach to tackle this?
Am I missing something obvious?
I have a vague idea that sorting may help, e.g. sort all mappings by the overall frequency of each descriptor by R position, but if my selection procedure is not good...
Any help would be appreciated.
Thanks!
L
I'll explain it as concisely as I can, but sorry, it will be a long post, it's not that trivial.
In chemistry there is an operation called 'R group decomposition' that is performed on a set of molecules to analyse the structural determinants of some of their properties.
Here's an example of template ('scaffold') that can be used:
There is some software that takes this template + a set of molecules, checks which ones 'match' the template and lists the corresponding R1, R2 and R3. These are the 'R groups', and taken together with the scaffold, uniquely identify a molecule. So far so good.
However, there are templates that, by effect of symmetry and chemical rules, can produce several valid sets of R groups. For instance this one:
Symmetry shows that R1,R2 can be swapped with R3,R4.
Chemistry shows that R1 and R2 can be swapped, and so can R3 and R4.
So, a molecule like this one:
has 8 'mappings':
M1_{R1,R2,R3,R4}={H,Me,Ph,Et},{H,Me,Et,Ph},{Me,H,Ph,Et},{Me,H,Et,Ph},{Ph,Et,H,Me},{Ph,Et,Me,H},{Et,Ph,H,Me},{Et,Ph,Me,H}.
Each of these mappings is perfectly valid. If one were only interested in identifying molecules, choosing anyone mapping at random would be OK.
The problem is that, when analysing the properties of a set of molecules, it is desirable to minimise the number of distinct 'descriptors' at each R-position and maximise the sum of all pairwise differences of numbers of descriptors; choosing mappings at random does not achieve that.
Example: suppose that we choose the first mapping for M1:
M1_{R1,R2,R3,R4}={H,Me,Ph,Et}
In terms of descriptors at each R position, we have: {R1}={H}, {R2}={Me}, {R3}={Ph}, {R4}={Et}.
Now we add this molecule to the set:
Due to the identity between some groups, the non-degenerate mappings are fewer:
M2_{R1,R2,R3,R4}={H,Me,H,Me},{H,Me,Me,H},{Me,H,Me,H},{Me,H,H,Me}.
It is inevitable to add new descriptors to 'some' R positions.
If we choose the first mapping of M2, we're doing well, because we don't add any new descriptor to R1 or R2, and we're adding new descriptors only to R3 and R4.
M2_{R1,R2,R3,R4}={H,Me,H,Me}
And we have now: {R1}={H}, {R2}={Me}, {R3}={Ph,H}, {R4}={Et,Me}.
The number of descriptors in each R position is just the number of distinct elements in each of these sets (so 1, 1, 2, 2 here). The total number of descriptors is the sum of the above, so here 1+1+2+2=6 (you don't consider equal the H in R1 and the H in R3, because at this point they are desymmetrised.
Obviously if we had chosen the 3rd mapping for M2, we would have added one new descriptor to each position, ignoring the fact that H was already in R1 and Me in R2.
It may seem simple: just choose for each new molecule the mapping that adds the fewest descriptors.
But it's not. First, you don't really add the molecules one by one: you already have a large set of molecules (e.g. 500) with all their combinatorial mappings (could be from 2 to 8, typically, for each molecule), and in the end you need to choose one mapping per molecule in such a way that the above conditions are satisfied (minimal sum of different descriptors, maximal sum of differences of numbers of descriptors). I don't need to tell you how many possible combinations that would make, if one wanted to approach this by brute force :O)
So I went for what I thought was a more 'clever' approach.
I tried this: pick one arbitrary mapping for the first molecule and then run iterations among the rest of the set to choose at each cycle one mapping that gives the lowest addition of descriptors overall, and within this condition, adds the new descriptors preferably to groups that already have more descriptors.
Computationally, I just made a vector like {1,0,0,1} for each mapping, comparing existing descriptors with the one in the mapping, 1 for not present, 0 for already present. Then I made a SUM composed by the norm of the vector (in this case 2) multiplied by 104, + the individual vector components each multiplied by 10i, where i is the appropriately chosen exponent from 0 to 3, based on the sorting by descriptor counts in the current reference sets of descriptors in the R positions. So you have 10001 for a mapping that adds 1 new descriptor to the R position that already has the most descriptors. 10010 would indicate adding 1 new descriptor to the R position that has the second most descriptors, and so on, 20011 would add 1 to each the most populated R position and the second most populated one. So at each cycle only mappings with the lowest SUM are considered, and only one of them is selected (because two mappings with SUM 10001 don't necessarily add the same descriptor to that position). The corresponding molecule passes, the reference R position sets of descriptors are updated, all other molecules are sent to the next iteration.
This seemed to work to some extent, but by running some tests I found that the initial choice, and even the order of the molecules, resulted in different end sets, some with a good separation of descriptors, some less good, and even the total number of descriptors varied a bit.
Example: in one run I got something like 13, 80, 220, 262; in another run something like 40,65,250,230.
This is not good. The solution should not only be unique, but also optimal. I'm not sure it can be independent from the initial choice of mapping for the first compound, but then there should be a way to choose that best.
Here's an example set:
M1 ={H,Me,Ph,Et},{H,Me,Et,Ph},{Me,H,Ph,Et},{Me,H,Et,Ph},{Ph,Et,H,Me},{Ph,Et,Me,H},{Et,Ph,H,Me},{Et,Ph,Me,H}
M2 ={H,Me,H,Me},{H,Me,Me,H},{Me,H,Me,H},{Me,H,H,Me}
M3={H,H,Me,Me},{Me,Me,H,H}
Can anyone please suggest an approach to tackle this?
Am I missing something obvious?
I have a vague idea that sorting may help, e.g. sort all mappings by the overall frequency of each descriptor by R position, but if my selection procedure is not good...
Any help would be appreciated.
Thanks!
L
Last edited: