- #1
query_ious
- 23
- 0
I'd appreciate advice on the correct statistical method to analyse a dataset -
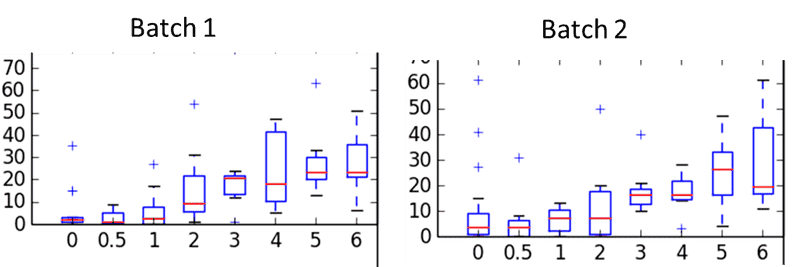
Dataset is basically a titration curve consisting of [0.5, 1, 2, 3, 4, 5, 6] pg of starting material and 8 replicates in each 'pg bin'. In 'stage 1' of the process each bin is labeled separately, in 'stage 2' all bins are pooled into one tube and amplified together and in 'stage 3' this is thrown into a DNA sequencer and bins are separated based on the labels.
So: the original input is a defined quantity of material and the final output is a number and what I would like to know is how accurately the output reflects the input. This feels like some kind of regression but I'm not sure what to use. The mean of 8 replicates? Their median? Something to do with the variability between them? Something based on a binomial sampling (with different bins having a different expected value?)?
Below is an example dataset - the x-axis is the titration curve bins ('0 pg' is background process noise), the y-axis is the 'output number' and the barplots are over the 8 replicates. There are 2 batches which are basically a repetition of the same experiment.
Thanks (and apologies if this has been asked before / is inappropriate)
<edit: uploading the image didn't work, I'm trying to fix this
for now - http://imgur.com/VRs5r6R
Dataset is basically a titration curve consisting of [0.5, 1, 2, 3, 4, 5, 6] pg of starting material and 8 replicates in each 'pg bin'. In 'stage 1' of the process each bin is labeled separately, in 'stage 2' all bins are pooled into one tube and amplified together and in 'stage 3' this is thrown into a DNA sequencer and bins are separated based on the labels.
So: the original input is a defined quantity of material and the final output is a number and what I would like to know is how accurately the output reflects the input. This feels like some kind of regression but I'm not sure what to use. The mean of 8 replicates? Their median? Something to do with the variability between them? Something based on a binomial sampling (with different bins having a different expected value?)?
Below is an example dataset - the x-axis is the titration curve bins ('0 pg' is background process noise), the y-axis is the 'output number' and the barplots are over the 8 replicates. There are 2 batches which are basically a repetition of the same experiment.
Thanks (and apologies if this has been asked before / is inappropriate)
<edit: uploading the image didn't work, I'm trying to fix this
for now - http://imgur.com/VRs5r6R
Last edited by a moderator: