- #1
rwooduk
- 762
- 59
Hi, maybe someone here could help explain this, I understand the concept of a nanopore; a graphene sheet with a hole burned in it, force DNA through it and measure the conductance to determine the base. But I'm unclear how micron sized beads help the process. Please find attached a slide from my lecture notes, any explanation of how the beads help sequencing, or what they are doing to the DNA would be really appreciated. The notes say the optical tweezers (that keep the bead in place) can manupulate the DNA, but why is there 3 strands of DNA attached to the bead? what is happening?
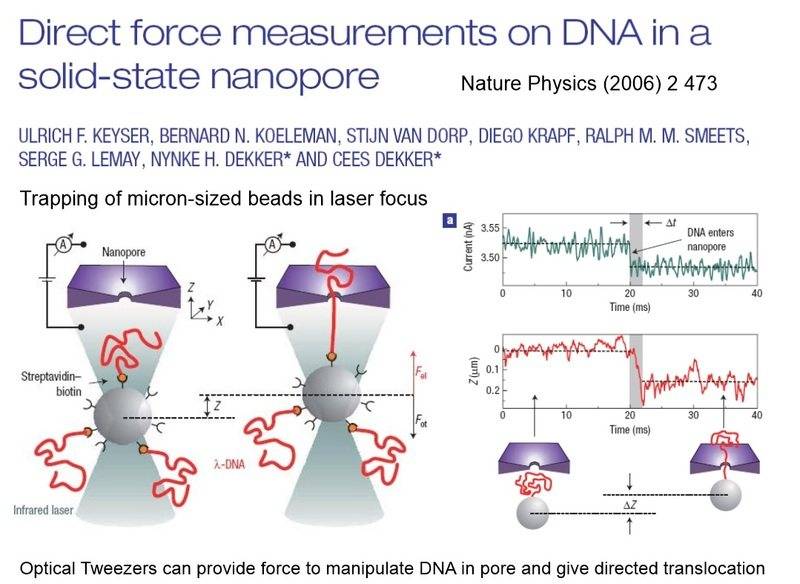
thanks for any help
thanks for any help