- #1
Kristoffer Linder
- 3
- 0
I am trying to make a deconvolution of fluorescence spectra in Matlab. The original spectra is the yellow graph in the figure below. The other two
graphs are Cauchy distributions, x and y, that I have manually added to the plot. I would like to write a program that could do this automatically for me, I have already tried deconv() but I do not get it to work, I have also try to estimate Gaussian distributions but it seems to me that the Cauchy distribution is more likely. I have tried to use conv(x,y,'same') to check if the manually found curved could be a used as an estimation, but it seems to me that after convolution I do not really get the desired answer, since I want an asymmetric curve, just as in the original graph. I need the component curves to be able to analyse the spectra correctly, since using the maxima of the original graph does not correspond very well with references. My Matlab code for the manual finding is
Anyone knows a good way to find the components of the original graph, and maybe what distribution to use? Also when using conv() in Matlab I get a problem with the scaling, since the output has much larger amplitude then the input.
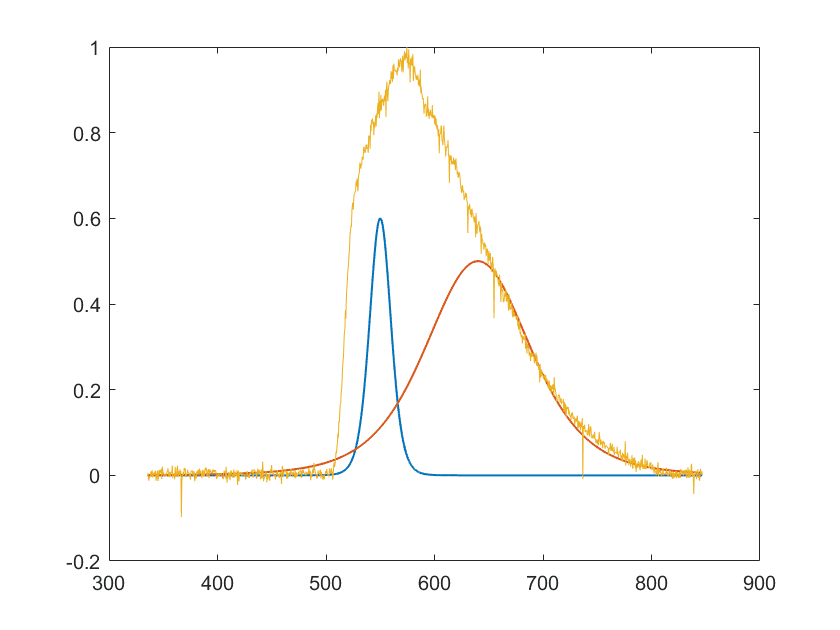
graphs are Cauchy distributions, x and y, that I have manually added to the plot. I would like to write a program that could do this automatically for me, I have already tried deconv() but I do not get it to work, I have also try to estimate Gaussian distributions but it seems to me that the Cauchy distribution is more likely. I have tried to use conv(x,y,'same') to check if the manually found curved could be a used as an estimation, but it seems to me that after convolution I do not really get the desired answer, since I want an asymmetric curve, just as in the original graph. I need the component curves to be able to analyse the spectra correctly, since using the maxima of the original graph does not correspond very well with references. My Matlab code for the manual finding is
Matlab:
close all
mu1 = 550;
mu2 = 640;
sigma1 = 10;
sigma2 = 50;
pd1 = makedist('tLocationScale','mu',mu1,'sigma',sigma1,'nu',9)
pd2 = makedist('tLocationScale','mu',mu2,'sigma',sigma2,'nu',5)
y1 = pdf(pd1,wavelength(100:1300,36));
y2 = pdf(pd2,wavelength(100:1300,36));
y1 = .6*y1/max(y1);
y2 = .5*y2/max(y2);
figure, plot(wavelength(100:1300,36),y1,'LineWidth',1)
hold on, plot(wavelength(100:1300,36),y2,'LineWidth',1)
hold on, plot(wavelength(100:1300,36),intensity(100:1300,36)/max(intensity(100:1300,36)))
Last edited by a moderator: