roam
- 1,265
- 12
I am trying to distinguish two different plant species based on their chemical signature using statistical signal classification. This is what the average curves of two species look like with their standard deviation bounds:
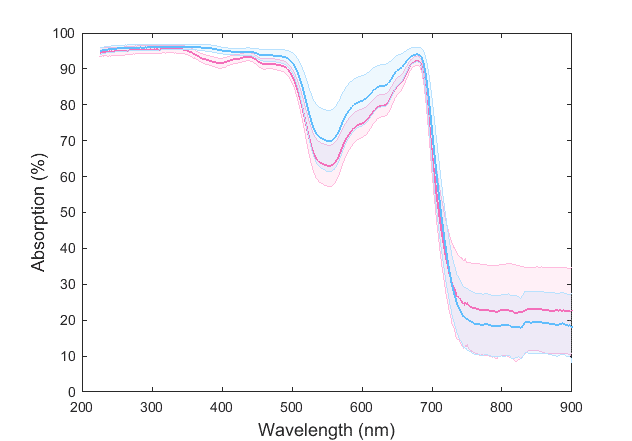
These spectra are made up of ~1400 points. So it was suggested to me to use a dimension reduction technique, instead of using a ~1400-dimensional vector space.
So, what are the problem that could occur if the multidimensional data space is large?
Also, if we were to classify based on only a few points (and throw away the rest) — would that not increase the likelihood of misclassification errors? Because we have fewer features to consider. I presume dimension reduction works differently because we are not throwing away any data.
Any explanation is greatly appreciated.
These spectra are made up of ~1400 points. So it was suggested to me to use a dimension reduction technique, instead of using a ~1400-dimensional vector space.
So, what are the problem that could occur if the multidimensional data space is large?
Also, if we were to classify based on only a few points (and throw away the rest) — would that not increase the likelihood of misclassification errors? Because we have fewer features to consider. I presume dimension reduction works differently because we are not throwing away any data.
Any explanation is greatly appreciated.
