- #1
roam
- 1,271
- 12
I am trying to use PCA to classify various spectra. I measured several samples to get an estimate of the population standard deviation (here I've shown only 7 measurements):
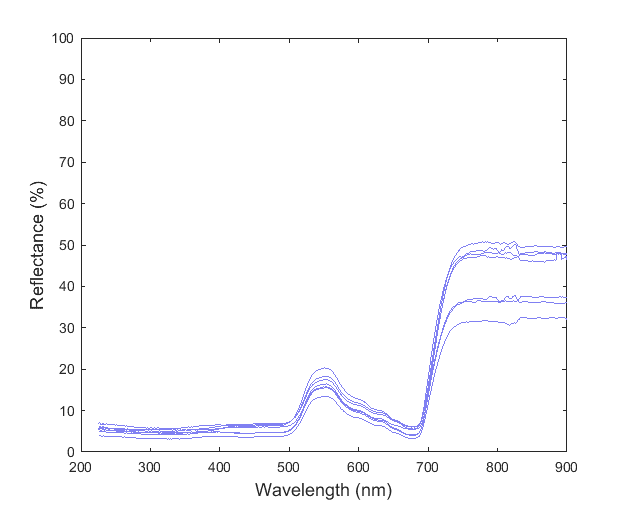
I combined all these data into a matrix where each measurement corresponded to a column. I then used the pca(...) function in Matlab to find the component coefficients. In this case, Matlab returned 6 components (not a significant dimension reduction since I had 7 measurements).
I plotted the first four sets of the component coefficients:
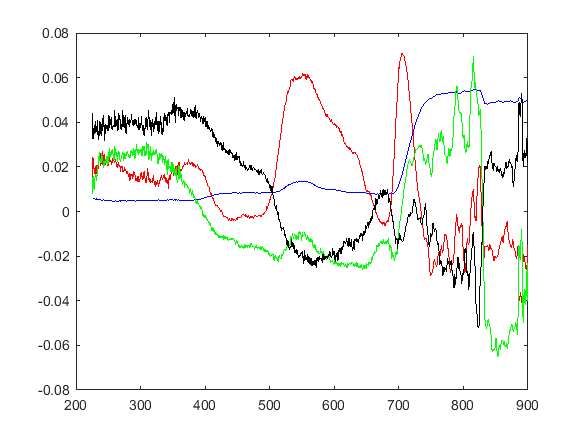
I am not sure how to interpret these curves. The blue curve is the first order coefficients and it resembles the overall shape of the measurement curves. The red curve is the second order coefficient set and it seems to accurately model the shape of the peak at 550 nm (but I don't understand the rest of the curve). The higher order coefficient sets were much noisier.
So, what do these curves represent exactly? Is it possible that each curve is influenced more by the presence of certain components (e.g. molecules of the substance that created the spectrum)?
I combined all these data into a matrix where each measurement corresponded to a column. I then used the pca(...) function in Matlab to find the component coefficients. In this case, Matlab returned 6 components (not a significant dimension reduction since I had 7 measurements).
I plotted the first four sets of the component coefficients:
I am not sure how to interpret these curves. The blue curve is the first order coefficients and it resembles the overall shape of the measurement curves. The red curve is the second order coefficient set and it seems to accurately model the shape of the peak at 550 nm (but I don't understand the rest of the curve). The higher order coefficient sets were much noisier.
So, what do these curves represent exactly? Is it possible that each curve is influenced more by the presence of certain components (e.g. molecules of the substance that created the spectrum)?





