- #1
fab13
- 312
- 6
I am currently studying Fisher's formalism as part of parameter estimation.
From this documentation :
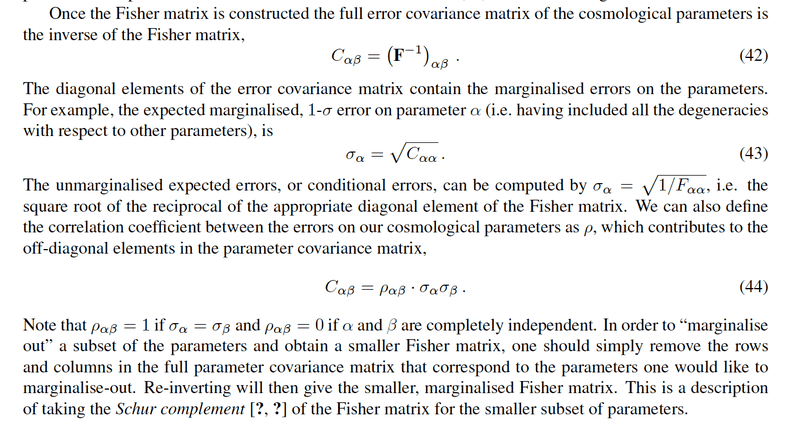
They that Fisher matrix is the inverse matrix of the covariance matrix. Initially, one builds a matrix "full" that takes into account all the parameters.
1) Projection : We can then do what we call a projection, that is to say that we express this matrix in another base by using the Jacobian matrix which involves the derivatives of the starting parameters with respect to the new parameters that one chooses: this matrix is called "projected matrix".
2) Marginalization : Once we have this projected matrix, we can do another operation which is the "marginalization of a parameter": that is to say that we delete in the projected matrix the row and the column corresponding to this marginalized parameter.
Finally, once I have the projected matrix, I can invert it to know the covariance matrix associated with the parameters (the new ones).
Now, I would like if the 2 following sequences gives the same final matrix :
1st sequence :
1.1) Starting from the Fisher matrix "full"
1.2) Invert "full" fisher Matrix to get Covariance matrix
1.3) Marginalize on Covariance matrix with respect to a parameter (or even several but I am interested first only in one), that is to say to remove the column and line corresponding to the parameter that one wants to marginalize.
1.4) Invert new Covariance matrix to get new Fisher
1.5) Project the new Fisher into new basis of parameters making the product: ## F_{\kappa \lambda} = \sum_ {i,j} \, J_{i \kappa} \, F_{ij} \, J _{\lambda j}##
2nd sequence :
2.1) Starting from the Fisher matrix "full"
2.2) Projecting with the Jacobian matrix
2.3) Invert to get new Covariance matrix
2.4) Marginalize the projected matrix = remove the column and line corresponding to the parameter that one wants to marginalize.
2.5) Invert to have the new Fisher matrix.
Will I get the same final Fisher matrix at the end of step 1.5) and step 2.5) ?
if this is the case, how could I prove it in an analytical way ?
Maybe the second sequence is not right to get the equivalence between sequence 1) and sequence 2) ?
From this documentation :
They that Fisher matrix is the inverse matrix of the covariance matrix. Initially, one builds a matrix "full" that takes into account all the parameters.
1) Projection : We can then do what we call a projection, that is to say that we express this matrix in another base by using the Jacobian matrix which involves the derivatives of the starting parameters with respect to the new parameters that one chooses: this matrix is called "projected matrix".
2) Marginalization : Once we have this projected matrix, we can do another operation which is the "marginalization of a parameter": that is to say that we delete in the projected matrix the row and the column corresponding to this marginalized parameter.
Finally, once I have the projected matrix, I can invert it to know the covariance matrix associated with the parameters (the new ones).
Now, I would like if the 2 following sequences gives the same final matrix :
1st sequence :
1.1) Starting from the Fisher matrix "full"
1.2) Invert "full" fisher Matrix to get Covariance matrix
1.3) Marginalize on Covariance matrix with respect to a parameter (or even several but I am interested first only in one), that is to say to remove the column and line corresponding to the parameter that one wants to marginalize.
1.4) Invert new Covariance matrix to get new Fisher
1.5) Project the new Fisher into new basis of parameters making the product: ## F_{\kappa \lambda} = \sum_ {i,j} \, J_{i \kappa} \, F_{ij} \, J _{\lambda j}##
2nd sequence :
2.1) Starting from the Fisher matrix "full"
2.2) Projecting with the Jacobian matrix
2.3) Invert to get new Covariance matrix
2.4) Marginalize the projected matrix = remove the column and line corresponding to the parameter that one wants to marginalize.
2.5) Invert to have the new Fisher matrix.
Will I get the same final Fisher matrix at the end of step 1.5) and step 2.5) ?
if this is the case, how could I prove it in an analytical way ?
Maybe the second sequence is not right to get the equivalence between sequence 1) and sequence 2) ?
