- #1
John37309
- 102
- 0
Identifying proteins by structure
Can proteins be identified by their structural image alone?
What do i mean by that? Well i understand the basics of protein folding and protein structure. I have a basic understanding of primary, secondary and tertiary structure of DNA/RNA. But i want to understand these images better.
Lets take Myoglobin as an example; http://en.wikipedia.org/wiki/Myoglobin
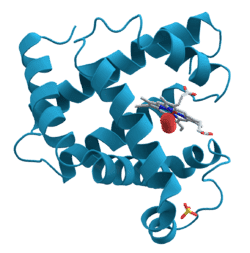
Quote Wikipedia;
" It has eight alpha helices and a hydrophobic core."
So can Myoglobin be identified by that picture alone? If i look at the picture, i can see 4 very large alpha helices and 4 smaller alpha helices. These are joined by short RNA strings.
In that Myoglobin image for example, the red alpha helices has 6 turns in the helix. Obviously if the red alpha helices had 10 turns instead of 6 turns, it would not be a Myoglobin protein. But my question is can proteins be identified just by counting the number of alpha helices and the number of turns in those individual alpha helices?
Is there any simple rule-of-thumb that you can use to allow you to identify different proteins just from the images?
John.
Can proteins be identified by their structural image alone?
What do i mean by that? Well i understand the basics of protein folding and protein structure. I have a basic understanding of primary, secondary and tertiary structure of DNA/RNA. But i want to understand these images better.
Lets take Myoglobin as an example; http://en.wikipedia.org/wiki/Myoglobin
Quote Wikipedia;
" It has eight alpha helices and a hydrophobic core."
So can Myoglobin be identified by that picture alone? If i look at the picture, i can see 4 very large alpha helices and 4 smaller alpha helices. These are joined by short RNA strings.
In that Myoglobin image for example, the red alpha helices has 6 turns in the helix. Obviously if the red alpha helices had 10 turns instead of 6 turns, it would not be a Myoglobin protein. But my question is can proteins be identified just by counting the number of alpha helices and the number of turns in those individual alpha helices?
Is there any simple rule-of-thumb that you can use to allow you to identify different proteins just from the images?
John.