- #1
roam
- 1,271
- 12
In the textbook “Principal Component Analysis” Jolliffe (§9.2) suggests the following method for variable reduction:
“When the variables fall into well-defined clusters, there will be one high-variance PC and, except in the case of 'single-variable' clusters, one or more low-variance PCs associated with each cluster of variables. Thus, PCA will identify the presence of clusters among the variables, and can be thought of as a competitor to standard cluster analysis of variables. [...] Identifying clusters of variables may be of general interest in investigating the structure of a data set but, more specifically, if we wish to reduce the number of variables without sacrificing too much information, then we could retain one variable from each cluster.”
I am trying to apply this idea of variable reduction to a problem which involves differentiating two substances based on their spectra which is composed of 1350 variables (i.e. wavelengths):
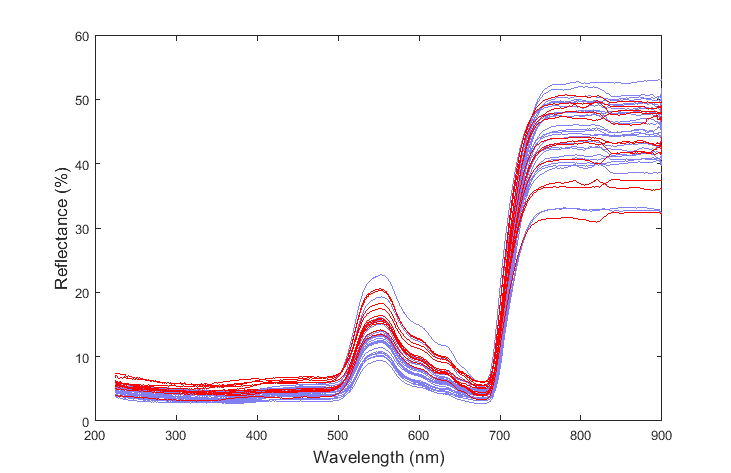
And here is the resulting differentiation between the species:
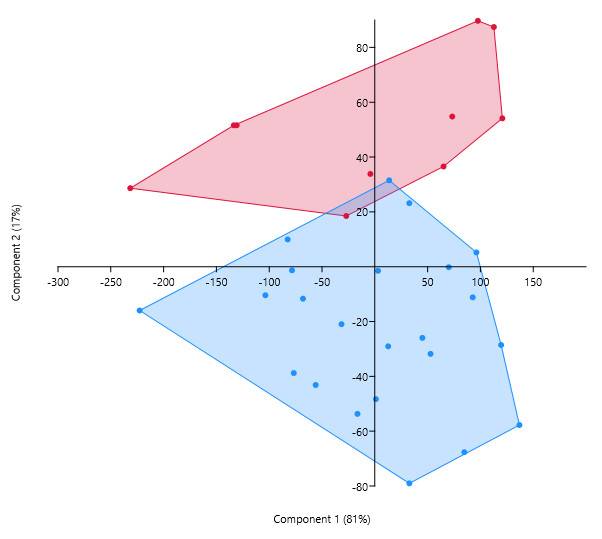
I want to be able to perform the same differentiation, but using fewer variables. Unfortunately, the textbook does not give any examples of how to do this. The aim is to divide variables, rather than observations, into groups. So, how should we plot the variables in order to see the clusters?
For instance, I have tried to plot the loadings at each variable with respect to the first two PCs and obtained:
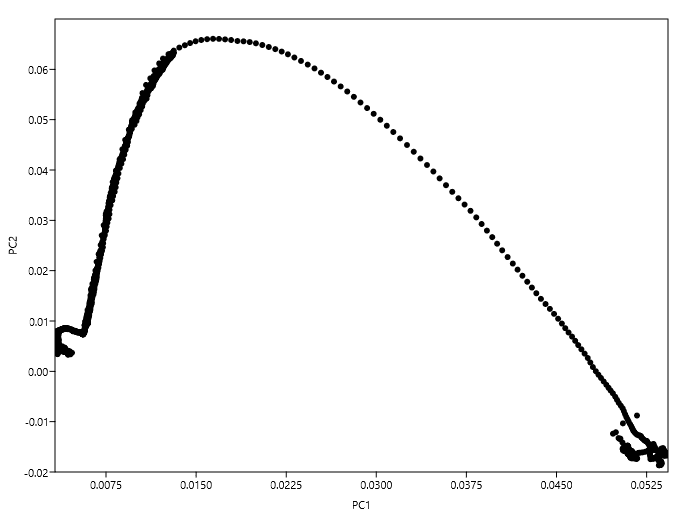
In this plot there doesn't seem to be any well-defined clusters to choose variables from. Am I following Jolliffe's method correctly?
Any explanation would be greatly appreciated.
P.S. Plotting the actual PCs give the following (the ordinate should represent the correlation coefficients between a given variable and the PC):
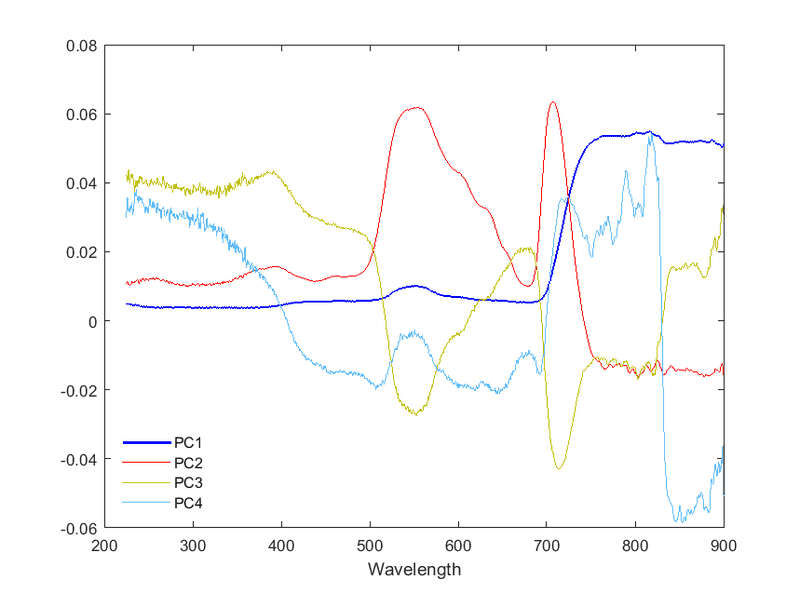
“When the variables fall into well-defined clusters, there will be one high-variance PC and, except in the case of 'single-variable' clusters, one or more low-variance PCs associated with each cluster of variables. Thus, PCA will identify the presence of clusters among the variables, and can be thought of as a competitor to standard cluster analysis of variables. [...] Identifying clusters of variables may be of general interest in investigating the structure of a data set but, more specifically, if we wish to reduce the number of variables without sacrificing too much information, then we could retain one variable from each cluster.”
I am trying to apply this idea of variable reduction to a problem which involves differentiating two substances based on their spectra which is composed of 1350 variables (i.e. wavelengths):
And here is the resulting differentiation between the species:
I want to be able to perform the same differentiation, but using fewer variables. Unfortunately, the textbook does not give any examples of how to do this. The aim is to divide variables, rather than observations, into groups. So, how should we plot the variables in order to see the clusters?
For instance, I have tried to plot the loadings at each variable with respect to the first two PCs and obtained:
In this plot there doesn't seem to be any well-defined clusters to choose variables from. Am I following Jolliffe's method correctly?
Any explanation would be greatly appreciated.
P.S. Plotting the actual PCs give the following (the ordinate should represent the correlation coefficients between a given variable and the PC):




