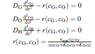clear all;
numx =50; %number of grid points in space
numt =82000; %number of time steps to be iterated over
tmax = .06;
Length =50E-7; %length of grid
DG = 4E-10; %requirement DG(dt)/dx^2 < .5, cm^2/sec
DO = 5E-9; %requirement DO(dt)/dx^2 < .5, cm^2/sec
DH = 5E-9; %requirement DH(dt)/dx^2 < .5, cm^2/sec
Vmax = 60E-6; %A/cm^2
KG = 33E-6; %mol/cm^3
KO = .2E-6;
G0 =2E-6; %mol/cm^3 initial substrate concentration
O20 = .12E-6; %mol/cm^3 inital oxygen concentration
cG = zeros(numx,1); %initialize everything to zero
cH = zeros(numx,1); %initialize everything to zero
cO = zeros(numx,1);
zcG = linspace(0,Length,numx)'; %vector of x values, to be used for plotting
tG = linspace(0,tmax,numt)'; %vector of t values, to be used for plotting
zcO = linspace(0,Length,numx)'; %vector of x values, to be used for plotting
tO = linspace(0,tmax,numt)'; %vector of t values, to be used for plotting
zcH = linspace(0,Length,numx)'; %vector of x values, to be used for plotting
tH = linspace(0,tmax,numt)'; %vector of t values, to be used for plotting
dxG = zcG(2)-zcG(1); %Define grid spacing in time
dtG = tG(2)-tG(1); %Define grid spacing in time
dxO = zcO(2)-zcO(1); %Define grid spacing in time
dtO = tO(2)-tO(1); %Define grid spacing in time
dxH = zcH(2)-zcH(1); %Define grid spacing in time
dtH = tH(2)-tH(1); %Define grid spacing in time
S_Stability1 = (DG*dtG)/((dxG)^2) %check requirement DG(dtG)/dxG^2 < .5, cm^2/sec
O_Stability1 = (DO*dtO)/((dxO)^2) %check requirement DO(dtO)/dxO^2 < .5, cm^2/sec
H_Stability1 = (DH*dtH)/((dxH)^2) %check requirement DH(dtH)/dxH^2 < .5
%{
Dirchelet Boundary Conditions, where Neumann BC dcG/dx=0 and dcO/dx=0
is in body of for loop below, IC is given by matrix initialization (zeros) and BC
%}
cG(numx) = G0; %cG(x=d,t)=G0
cO(numx) = O20; %cO(x=d,t)=O20
cH(1) = 0; %cH(x=0,t)=0
cH(numx) = 0; %cH(x=d,t)=0
%iterate central difference equation%
for j=1:numt %time loop
cGlast = cG;
cOlast = cO;
cHlast = cH;
for i=2:numx-1 %space loop
cG(i) = cGlast(i) + (dtG/dxG^2)*DG*(cGlast(i+1) - 2*cGlast(i) + cGlast(i-1))-((Vmax*dtG*cGlast(i)*cOlast(i))/(cGlast(i)*cOlast(i)+(KG*cOlast(i))+KO*cGlast(i)));
cO(i) = cOlast(i) + (dtO/dxO^2)*DO*(cOlast(i+1) - 2*cOlast(i) + cOlast(i-1))-((Vmax*dtO*cGlast(i)*cOlast(i))/(cGlast(i)*cOlast(i)+(KG*cOlast(i))+KO*cGlast(i)));
cH(i) = cHlast(i) + (dtH/dxH^2)*DH*(cHlast(i+1) - 2*cHlast(i) + cHlast(i-1))+((Vmax*dtH*cGlast(i)*cOlast(i))/(cGlast(i)*cOlast(i)+(KG*cOlast(i))+KO*cGlast(i)));
y(j,1) = cH(2); %store data in vector for current plotting
end
cG(1)= cGlast(1)+dtG*DG*2*(cGlast(2)-cGlast(1))./dxG.^2; %Neumann Boundary Condition
cO(1)= cOlast(1)+dtO*DO*2*(cOlast(2)-cOlast(1))./dxO.^2; %Neumann Boundary Condition
end
figure(1);
plot(zcH,cH);
xlabel('Length (cm)');
ylabel('[H2O2] (M)');
figure(2);
plot(zcG,cG);
xlabel('Length (cm)');
ylabel('[Glucose] (M)');
figure(3);
plot(zcO,cO);
xlabel('Length (cm)');
ylabel('[O2] (M)');
figure(4);
i = 2*(96500)*DH*(y/dxH);
%i = 2*(96500)*DH*(y/dxH);
plot(tH,i);
xlabel('Time (s)');
ylabel('Current (A)');