- #1
user366312
Gold Member
- 89
- 3
- TL;DR Summary
- Suppose I have a protein PDB file and want to compute energy using the following formula from the coordinates in the file.
**How can I do that?**
Suppose I have a protein PDB file and want to compute energy using the following formula from the coordinates in that file.
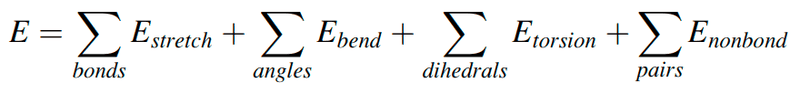
How can I do that?
Sometimes people suggest using the `CONECT` field for that purpose. However, some PDB files do not have this field.
Example: 4OSK.pdb (Crystal structure of TAL effector reveals the recognition between asparagine and guanine)
How can I do that?
Sometimes people suggest using the `CONECT` field for that purpose. However, some PDB files do not have this field.
Example: 4OSK.pdb (Crystal structure of TAL effector reveals the recognition between asparagine and guanine)