- #1
bcolson
- 3
- 0
I have been having trouble getting the calculation of energy for a chain of coupled oscillators to come out correctly. The program was run in Matlab and is intended to calculate the energy of a system of connected Hooke's law oscillators. Right now there is only stiffness and no dampening effects or force functions.
Half of the system has stiffness 1 the other half has variable stiffness.
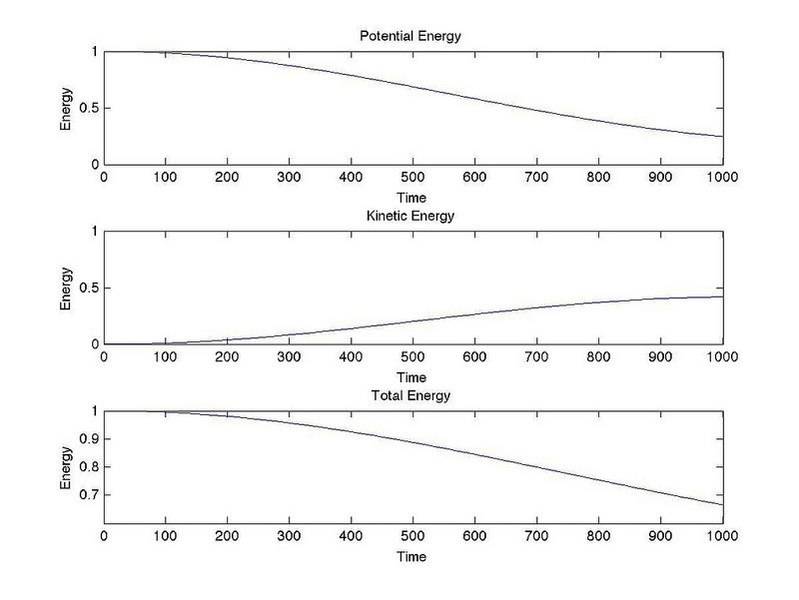
When I ran the program to calculate the energy of a system where stiffness = 1, 4 total masses, no kinetic energy or potential energy except on mass #1 and plotted out the energy vs time a large chunk of energy disappeared.
 the definitions of the variables are:
the definitions of the variables are:
positionMatrix == a matrix of positions generated by another program
velocityMatrix == a matrix of velocities generated by yet another program
stiffness == the stiffness of the second half of the system
width == number of oscillating points
size == total number of steps
pointPotentialEnergy == the potential energy of each point at each step
pointKineticEnergy == the kinetic energy of each point at each step
totalPotentialEnergy == the total potential energy of the system at each step
totalKineticEnergy == the total potential energy of the system at each step
This section of code calculates the energy, the problem is likely here.
I've shown the code the the group I am working with and they couldn't find any problems with the math. I strongly suspect that the problem is here though I can upload the other codes.
Half of the system has stiffness 1 the other half has variable stiffness.
When I ran the program to calculate the energy of a system where stiffness = 1, 4 total masses, no kinetic energy or potential energy except on mass #1 and plotted out the energy vs time a large chunk of energy disappeared.
positionMatrix == a matrix of positions generated by another program
velocityMatrix == a matrix of velocities generated by yet another program
stiffness == the stiffness of the second half of the system
width == number of oscillating points
size == total number of steps
pointPotentialEnergy == the potential energy of each point at each step
pointKineticEnergy == the kinetic energy of each point at each step
totalPotentialEnergy == the total potential energy of the system at each step
totalKineticEnergy == the total potential energy of the system at each step
This section of code calculates the energy, the problem is likely here.
Matlab:
function [pointPotentialEnergy,pointKineticEnergy,totalPotentialEnergy,totalKineticEnergy]=energyCalculator3(positionMatrix,velocityMatrix,stiffness)
%initialize vectors
width = length(positionMatrix(:,1));
size = length(positionMatrix(1,:));
pointPotentialEnergy = zeros(width,size);
pointKineticEnergy = zeros(width,size);
totalPotentialEnergy = zeros(size,1);
totalKineticEnergy = zeros(size,1);
for n = 1:size
for p = 1:width
pointKineticEnergy(p,n) = 0.5*velocityMatrix(p,n)^2;
if p == 1
pointPotentialEnergy(p,n) = 0.5*positionMatrix(p,n)*positionMatrix(p,n);
elseif p < (width*0.5)
pointPotentialEnergy(p,n) = 0.5*(positionMatrix(p,n)-positionMatrix(p-1,n))^2;
elseif p == (width*0.5)
pointPotentialEnergy(p,n) = 0.5*stiffness*(positionMatrix(p,n)-positionMatrix(p-1,n))^2;
elseif p == width
pointPotentialEnergy(p,n) = 0.5*stiffness*positionMatrix(p,n)*positionMatrix(p,n);
else
pointPotentialEnergy(p,n) = 0.5*stiffness*(positionMatrix(p,n)-positionMatrix(p-1,n))^2;
end
end
end
for q = 1:size
totalPotentialEnergy(q) = sum(pointPotentialEnergy(:,q));
totalKineticEnergy(q) = sum(pointKineticEnergy(:,q));
end