- #1
- 3,759
- 4,199
All life on Earth stores its genetic information in DNA using just four nucleotide letters: A, T, C, and G. Research published this week in the journal Nature describes how scientists engineered a bacterium to incorporate two new letters into their DNA (which they call X and Y, pictured below), and read those letters to introduce new amino acids into proteins.
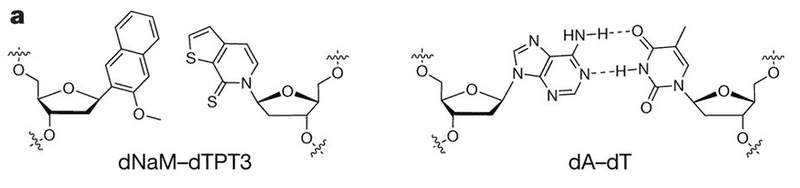
(the X-Y base pair is shown on the left. A natural A-T base pair is shown for comparision on the right)
The work builds upon previous work by the same group. In 2014, the team reported that they could get bacteria to incorporate an unnatural base pair into DNA and RNA, and earlier this year, they reported improvements to enable the bacteria to more stably maintain the unnatural base pair in their DNA. Of course, while getting an unnatural amino acid into DNA and RNA is in itself an important achievement, the authors were missing the last step of biology. DNA gets transcribed into mRNA which then gets read by the ribosome to produce protein. While the bacteria contained the extra base pair, their ribosomes lacked the capability to read the new nucleotides.
The latest work involves engineering the tRNAs and related machinery so that the ribosome can recognize X in mRNA and incorporate unnatural amino acids into the resulting protein. Thus, these bacteria are now capable of containing extra information in their DNA and translating that information into an expanded genetic code that can encode more than the 20 amino acids typically made by life on Earth. It is worth noting, however, that they are many (much simpler!) approaches to introducing unnatural amino acids into protein. Furthermore, more work would need likely to be done for the bacteria to make more extensive use of the unnatural nucleotides and amino acids. Still, the work presents the first step towards potentially building new life forms in the lab that have an entirely different genetic system than any other life on Earth.
You can read the full paper here: https://www.nature.com/articles/nature24659
Popular press coverage:
https://www.npr.org/sections/health...s-move-a-step-closer-to-making-synthetic-life
https://www.technologyreview.com/s/609567/semi-synthetic-life-form-now-fully-armed-and-operational/
(the X-Y base pair is shown on the left. A natural A-T base pair is shown for comparision on the right)
The work builds upon previous work by the same group. In 2014, the team reported that they could get bacteria to incorporate an unnatural base pair into DNA and RNA, and earlier this year, they reported improvements to enable the bacteria to more stably maintain the unnatural base pair in their DNA. Of course, while getting an unnatural amino acid into DNA and RNA is in itself an important achievement, the authors were missing the last step of biology. DNA gets transcribed into mRNA which then gets read by the ribosome to produce protein. While the bacteria contained the extra base pair, their ribosomes lacked the capability to read the new nucleotides.
The latest work involves engineering the tRNAs and related machinery so that the ribosome can recognize X in mRNA and incorporate unnatural amino acids into the resulting protein. Thus, these bacteria are now capable of containing extra information in their DNA and translating that information into an expanded genetic code that can encode more than the 20 amino acids typically made by life on Earth. It is worth noting, however, that they are many (much simpler!) approaches to introducing unnatural amino acids into protein. Furthermore, more work would need likely to be done for the bacteria to make more extensive use of the unnatural nucleotides and amino acids. Still, the work presents the first step towards potentially building new life forms in the lab that have an entirely different genetic system than any other life on Earth.
You can read the full paper here: https://www.nature.com/articles/nature24659
Popular press coverage:
https://www.npr.org/sections/health...s-move-a-step-closer-to-making-synthetic-life
https://www.technologyreview.com/s/609567/semi-synthetic-life-form-now-fully-armed-and-operational/
Attachments
Last edited: