- #1
- 2,585
- 10,560
- TL;DR Summary
- References to something new in biology, with links and short descriptions of why its interesting.
May not be open access.
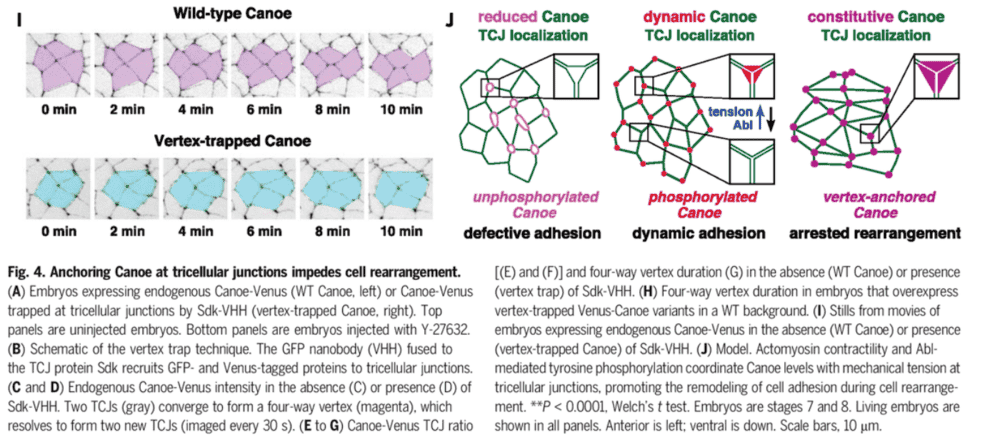
Molecular regulators of inter-cellular tension, at triple junctions between cells, in an epithelium have been described.
Probably behind paywall.
Interpretation by non-authors.
Research article.
An epithelium is a kind of tissue, where the cells are arranged in sheets.
These cellular sheets have two different sides: inside/outside (basal/apical).
Different structures form inside the cells at the different ends. The cells are polarized in that direction.
This is probably one of the most phylogenetically ancient kinds of metazoan tissues.
It is involved in defining the difference between the organism and its environment, and containing its controlled internal environment.
Epithelia are held together, in sheets, by special contacts between the cells made of proteins that also interact across the membrane (through interactions involving other proteins) with the cytoskeleton (internal structural members of the cell).
This article is about the dynamic cellular control of these intercellular biophysical forces.

This kind of thing was probably well evolved in the early metazoans (multicellular) animals that had three tissue layers : endoderm (internal epithelia, for digestive purposes), ectoderm (external epilethial (for maintaning internal environment, repelling parasites), and mesoderm (usually non-epithelial, lots of tissues between the inner and outer epithelia).
Each new tissue type would have to evolve a bunch of new mechanisms, encoded in genes and their controls.
This little mechanism seems to involve at least two novel proteins.
It is probably widely inherited among the metazoans (multicellular animals).
This is why it is interesting to me. Another step in the evolution of greater organismal complexity by changing a few things in a large genome.
Probably behind paywall.
Interpretation by non-authors.
Research article.
An epithelium is a kind of tissue, where the cells are arranged in sheets.
These cellular sheets have two different sides: inside/outside (basal/apical).
Different structures form inside the cells at the different ends. The cells are polarized in that direction.
This is probably one of the most phylogenetically ancient kinds of metazoan tissues.
It is involved in defining the difference between the organism and its environment, and containing its controlled internal environment.
Epithelia are held together, in sheets, by special contacts between the cells made of proteins that also interact across the membrane (through interactions involving other proteins) with the cytoskeleton (internal structural members of the cell).
This article is about the dynamic cellular control of these intercellular biophysical forces.
This kind of thing was probably well evolved in the early metazoans (multicellular) animals that had three tissue layers : endoderm (internal epithelia, for digestive purposes), ectoderm (external epilethial (for maintaning internal environment, repelling parasites), and mesoderm (usually non-epithelial, lots of tissues between the inner and outer epithelia).
Each new tissue type would have to evolve a bunch of new mechanisms, encoded in genes and their controls.
This little mechanism seems to involve at least two novel proteins.
It is probably widely inherited among the metazoans (multicellular animals).
This is why it is interesting to me. Another step in the evolution of greater organismal complexity by changing a few things in a large genome.