- #1
fog37
- 1,568
- 108
- TL;DR Summary
- Docker base image...
Hello,
anyone on Physics Forums using Docker? I have understood the overarching idea behind Docker (packaging code, dependencies, etc. into a container that can be shared across OSs) but I have some doubts:
I am confused about the base image line of code in the Dockerfile (which is simple text file): FROM python: 3.9
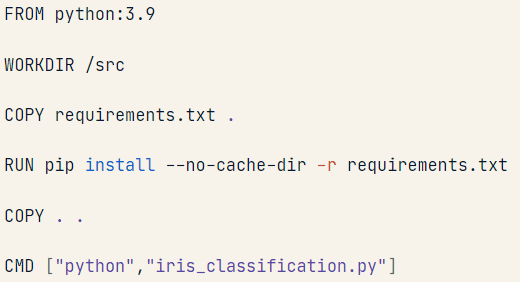
anyone on Physics Forums using Docker? I have understood the overarching idea behind Docker (packaging code, dependencies, etc. into a container that can be shared across OSs) but I have some doubts:
I am confused about the base image line of code in the Dockerfile (which is simple text file): FROM python: 3.9
- Why is that instruction called an "image"?
- That first line line specifies the Python interpreter to use inside the container...Does that mean that when the container is built, using the instructions in the Dockerfile, a python 3.9 interpreter gets also downloaded and used into the built container?
- Where does that specified interpreter get exactly downloaded from? When creating the docker container, the code and the dependencies are on my local machine....Is the Python interpreter instead downloaded form the internet?
- The last instruction, CMD ["python, iris_classification.py"], specifies what to do at the command line to launch the program, i.e. type python followed by the name of the script, correct? Would it be possible to launch the program as a Jupyter notebook (ipynb) or does the code need to be in a .py file?